Vina
What is Vina
AutoDock Vina is a software program widely used in computational biology for the purpose of drug discovery and molecular docking. It is particularly renowned for its speed, accuracy, and ease of use in predicting how small molecules, such as substrates or drug candidates, bind to a receptor of known 3D structure.
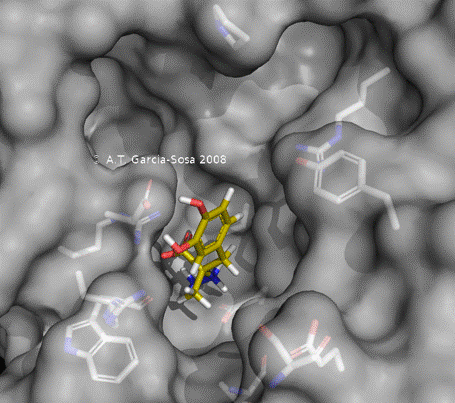
When working with AutoDock Vina, one common challenge that users may encounter is the time it takes to complete docking simulations, especially when dealing with large numbers of ligands or particularly complex receptor structures. Thus, hardware acceleration approaches of Vina are the main topics of Vina group. Current acceleration of AutoDock Vina typically relies on the stack of computing power as well as the allocation of resource and tasks.
Research
Vina group is making efforts to accelerate vina with GPU, FPGA and recyclable smartphones.
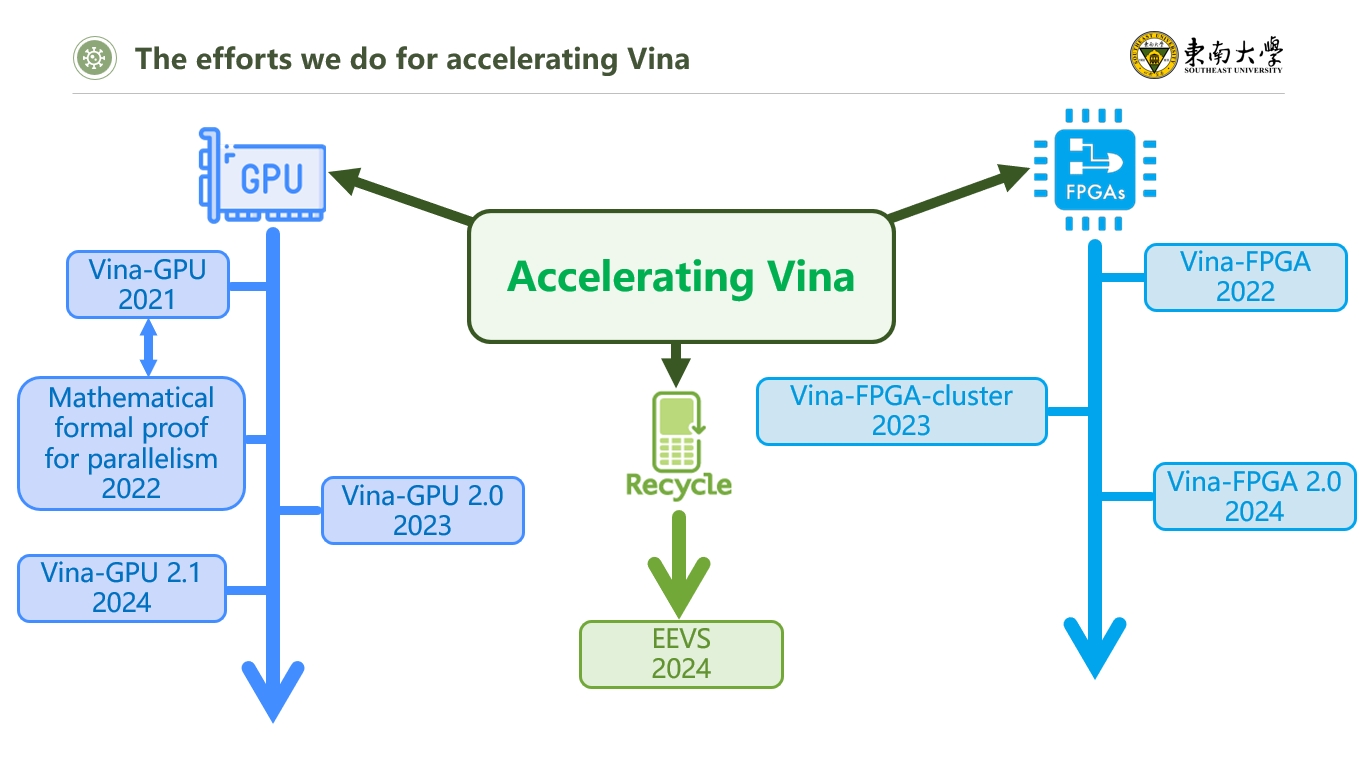
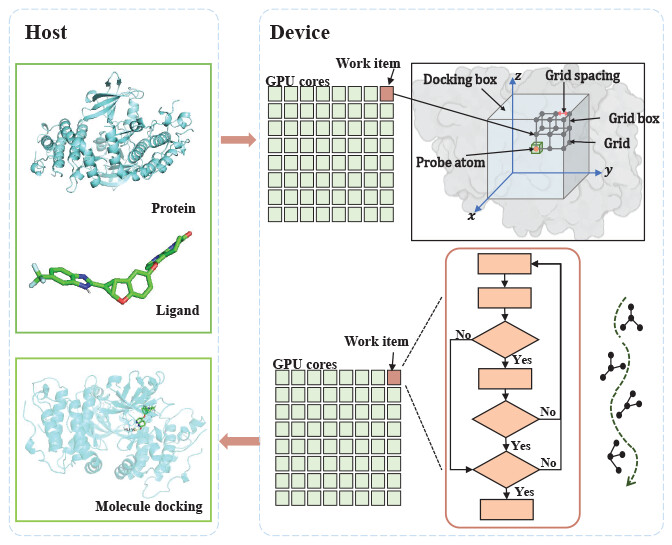
1. GPU
Our recent Vina-GPU method realized 14-fold acceleration against AutoDock Vina on a piece of NVIDIA RTX 3090 GPU in one virtual screening case. Further speedup of AutoDock Vina and its derivatives with graphics processing units (GPUs) is beneficial to systematically push their popularization in large-scale virtual screens due to their high benefit–cost ratio and easy operation for users. Thus, we proposed the Vina-GPU 2.0 method to further accelerate AutoDock Vina and the most common derivatives with new docking algorithms (QuickVina 2 and QuickVina-W) with GPUs.
Paper: 10.1021/acs.jcim.2c01504
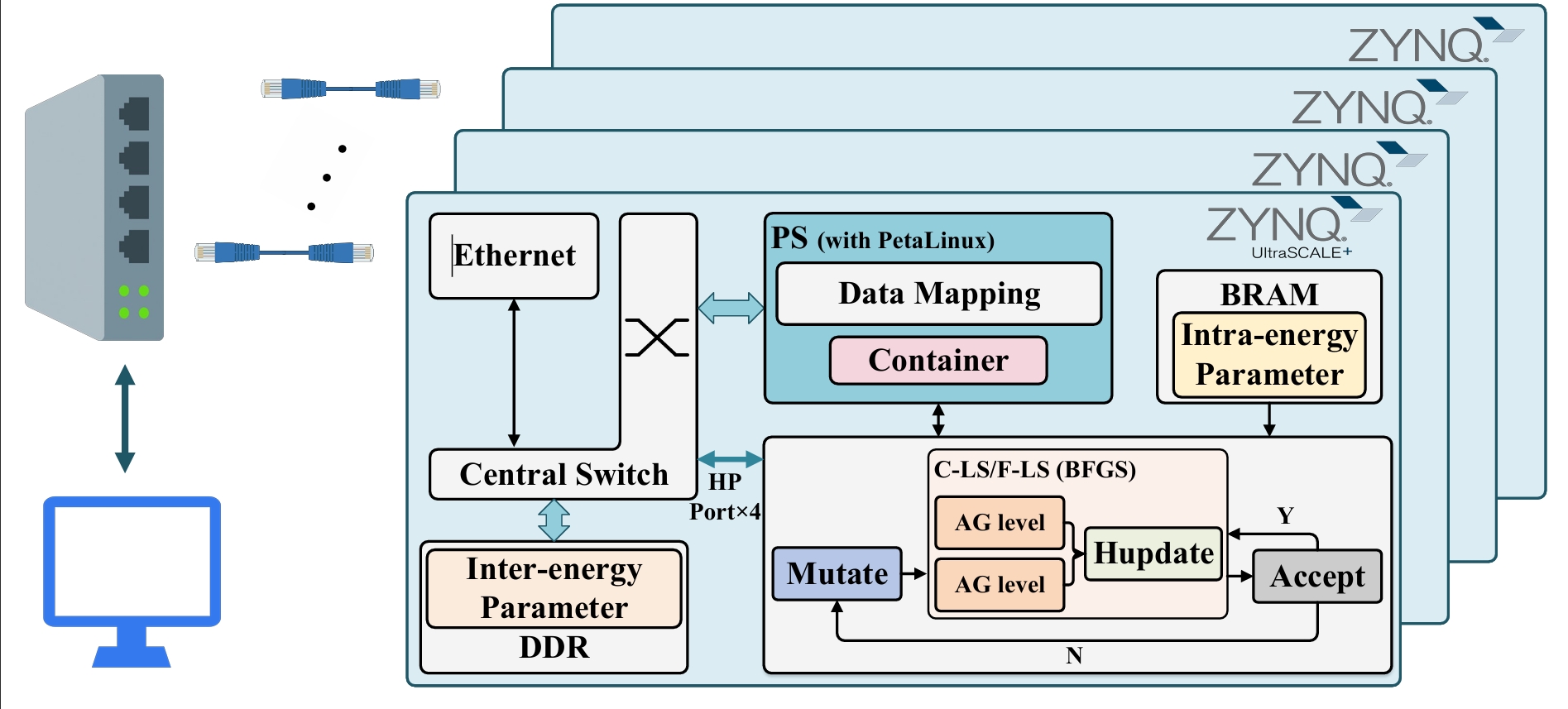
2. FPGA
Hardware acceleration of Vina on FPGA platforms offers a high energy-efficiency approach to speed up the docking process. we propose Vina-FPGA-cluster, a multi-FPGA-based molecular docking tool enabling high-accuracy and multi-level parallel Vina acceleration. Standing upon the shoulders of Vina-FPGA, we first adapt hybrid fixed-point quantization to minimize accuracy loss. We then propose a SystemC-based model, accelerating the hardware accelerator architecture design evaluation. Next, we propose a novel bidirectional AG module for data-level parallelism. Finally, we optimize the system architecture for scalable deployment on multiple Xilinx ZCU104 boards, achieving task-level parallelism.
3. Smartphones
Virtual screening plays an indispensable role in the early stages of drug discovery, which utilizes high-throughput molecular docking to find potential drug candidates from vast databases. This work introduces EEVS (Economical and Ecological Virtual Screening), an innovative framework that utilizes the computational capabilities of discarded smartphones for cost-effective and ecofriendly virtual screening. To the best of our knowledge, we are the first to propose a virtual screening framework that leverages discarded smartphones to accelerate drug discovery.

Publications
Vina-FPGA2: a high-level parallelized hardware-accelerated molecular docking tool based on the inter-module pipeline
- Ming Ling, Shidi Tang, Ruiqi Chen, Xin Li, Yanxiang Zhu
- Frontiers of Information Technology & Electronic Engineering
- DOI: 10.1631/FITEE.2400941
DiffDock-FPGA: A Hardware Accelerator for Molecular Docking with Customized Tensor Product Framework and Sparse-Aware Access Strategy
- Chuanzhao Zhang, Rui Liu, Shidi Tang, Shun Li, Ming Ling
- Proceedings of the Great Lakes Symposium on VLSI 2025 (GLSVLSI)
- DOI: 10.1145/3716368.3735212
EEVS: Redeploying Discarded Smartphones for Economic and Ecological Drug Molecules Virtual Screening
- Ming Ling, Chuanzhao Zhang, Shidi Tang, Ruiqi Chen, Yanxiang Zhu
- IEEE Transactions on Sustainable Computing
- DOI: 10.1109/TSUSC.2025.3541958
Vina-FPGA-Cluster: Multi-FPGA Based Molecular Docking Tool with High-Accuracy and Multi-Level Parallelism
- Ming Ling, Zhihao Feng, Ruiqi Chen, Yi Shao, Shidi Tang, Yanxiang Zhu
- 2024 IEEE Transactions on Biomedical Circuits and Systems (TBCAS)
- DOI: 10.1109/TBCAS.2024.3388323
Effectiveness Analysis of Multiple Initial States Simulated Annealing Algorithm, a Case Study on the Molecular Docking Tool Autodock Vina
- Xingxing Zhou, Ming Ling, Qingde Lin, Shidi Tang, Jiansheng Wu, Haifeng Hu
- 2023 IEEE/ACM Transactions on Computational Biology and Bioinformatics (TCBB)
- DOI: 10.1109/TCBB.2023.3323552
Vina-GPU 2.0: Further Accelerating AutoDock Vina and Its Derivatives with Graphics Processing Units
- Ji Ding, Shidi Tang, Zheming Mei, Lingyue Wang, Qinqin Huang, Haifeng Hu, Ming Ling, and Jiansheng Wu
- 2023 Journal of Chemical Information and Modeling (JCIM)
- DOI: 10.1021/acs.jcim.2c01504
Vina-FPGA: A Hardware-Accelerated Molecular Docking Tool With Fixed-Point Quantization and Low-Level Parallelism
- Ming Ling, Qingde Lin, Ruiqi Chen, Haimeng Qi, Mengru Lin, Yanxiang Zhu, Jiansheng Wu
- 2023 IEEE Transactions on Very Large Scale Integration (VLSI) Systems (TVLSI)
- DOI: 10.1109/TVLSI.2022.3217275
Accelerating autodock vina with gpus
- Shidi Tang, Ruiqi Chen, Mengru Lin, Qingde Lin, Yanxiang Zhu, Ji Ding, Haifeng Hu, Ming Ling, Jiansheng Wu
- 2022 Molecules
- DOI: 10.3390/molecules27093041
Latest edited in 2025.12
